В рамках планируемой коллаборации Казахстана и Китая по теме: «Ключ к здоровью: от генотипа к фенотипу» представляем разработки ученых из Фуданского университета ( Шанхай, КНР) и разработки Института генетики и физиологии КН МОН РК ( Казахстан)
Finding the «key» to health: from genome to phenotype
Academician of Chinese Academy of Sciences, President of Fudan University, Shanghai
Introduction
With the entry of life science research into the post-genome era, the «phenotype» has been regarded by the academic community as the next strategic commanding height in the field of life health after the «genome», and it is the «key» for further analysis of human health, which will serve as an important contribution to the cause of global health. It breeds a new scientific impetus and opens a new era of human exploration of the mysteries of life.
Gene sequencing and phenotypic measurements are the foundation of genetic research. With the help of the Human Genome Project, the field of life sciences has opened a new era of genetic research, and the systematic analysis of the relationship between genes and phenotypes has become a new frontier direction.
In 1911, W. Johannsen, a geneticist at the University of Copenhagen in Denmark, proposed the concept of «phenotype». In 1996, S. Garan at the University of California first proposed the concept of «phenomics». After years of exploration and accumulation of practice, the author of this paper’s research team defines phenome as a set of measurable characteristics resulting from the complex interactions between genes, epigenetics, commensal microorganisms, diet and environmental exposure, including Physical, chemical, and biological characteristics of individuals and groups [1].
Phenotypic groups are complex, cross-scale and dynamic. There is a complex regulatory network relationship between the genome and the phenotype, including single-gene regulation, pleiotropic-one-cause (that is, multiple genes regulate the same phenotype), and one-cause pleiotropic (that is, one gene regulates multiple phenotypes). At the same time, the phenotype group includes all phenotypes from microscopic to macroscopic, covering multiple levels such as transcription, protein, metabolism, cell, organ, and psychology. In addition, the phenotype group has the characteristics of dynamic changes with time and space that is, with the whole life cycle of the organism’s birth, growth and development, aging and death, as well as environmental changes such as altitude, temperature, and humidity, showing obvious dynamic changes.
As an emerging new interdisciplinary field, phenomics is dedicated to systematic phenotype research at the genome-wide level, and is an important engine in the field of biomedicine in the post-genome era [2].
Human Genome Project:
The first major international science program in the field of
life sciences
The Human Genome Project was completed ahead of schedule with the participation of scientists from six countries.
In the 1980s, American scientists gradually proposed to carry out human genome sequencing studies and phenotype association studies. In October 1990, the Human Genome Project (Human Genome Project) was officially launched, with the goal of accurately determining the human genome consisting of 3 billion base pairs. From 1996 to 1999, the United Kingdom, Germany, Japan, France and China successively joined the Human Genome Project and formed the International Genome Sequencing Consortium. China has undertaken the «1% Project» of the Human Genome Project, that is, the base sequencing task of chromosome 3. In June 2000, the draft human genome sequence was initially completed. In February 2001, we published the sequencing results of the human genome. In April 2003, the Human Genome Project, which has gathered the strength of international cooperation, was announced to be completed ahead of schedule [3].
Significance of the Human Genome Project
The Human Genome Project is a historic milestone for mankind to explore its own mysteries, bringing life science research into the era of genomes, promoting the continuous reduction of the cost of high-throughput gene sequencing, and promoting the development of research of proteomics and metabolomics, with breakthroughs on the molecular level for disease research and health management. As the first major international science project in the field of life sciences, the Human Genome Project covers scientific research, industry, ethics, society and other aspects, setting a model for the team building and management mechanism of international major scientific projects.
development of research of proteomics and metabolomics, with breakthroughs on the molecular level for disease research and health management. As the first major international science project in the field of life sciences, the Human Genome Project covers scientific research, industry, ethics, society and other aspects, setting a model for the team building and management mechanism of international major scientific projects.
As the only developing country participating in the Human Genome Project, China has made outstanding contributions in genome sequencing and genetic diversity. This has laid an important foundation for China to continue to participate in major international cooperation projects, such as the International HapMap Project (human genome haplotype map project), and to prepare for launching a major scientific project.
Although the Human Genome Project has greatly promoted the development of the life sciences, it is still very difficult to analyze the human health map and disease mechanisms jus based on genomic information. Taking psychiatric research as an example, scientists have been looking for stable and precise phenotypic indicators. However, screening valuable phenotypes and psychiatric identification criteria through genetic research is still full of challenges [4].
Carrying out human phenotype research has become the consensus of the scientific community in the post-genome era. After the completion of the Human Genome Project, life science research has entered a new era of large samples, big data, big science, and big discoveries. While scientists are focusing on genetic research under the light of big science, they are also looking forward to the arrival of the new era [5].
Classical genetics research usually has two strategies: one is the research strategy from phenotype to genotype, namely forward genetics, which focuses on research based on families or between different individuals; the other one is from genotype to phenotype, namely reverse genetics. It is usually limited by model organisms and it is difficult to use genetic editing technology to evaluate transgenic results on a large scale, especially because of ethical issues and cannot be carried out in human research [6]. However, phenome-wide research strategies, such as phenome-wide association studies, can help provide a new perspective for health and disease research [6].
Following the Human Genome Project, the importance of human phenotype research has attracted widespread attention, and the development of human phenotype research has become the consensus of the international academic community.
After the completion of the Human Genome Project, American scientists first proposed the term Human Phenome Project. However, hampered by the level of science and technology at that time, Americans based phenotype measurement and research mainly on medical electronic files and clinical laboratory tests..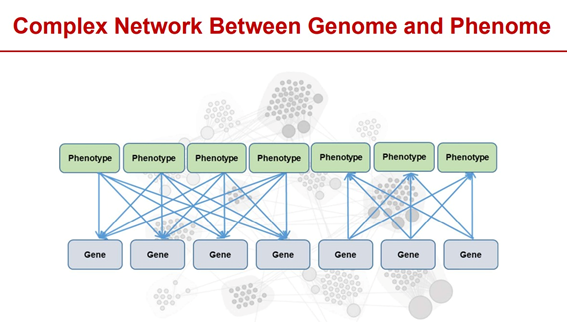
In 2012, the National Center for Biotechnology Information of the United States launched the construction of a public database (ClinVar) to conduct research on the relationship between human genetic variation and phenotype. In 2014, the Institute of Systems Biology of the United States launched the Hundred Person Wellness Project, which is a systematic study on the continuous monitoring of sleep and physiological activities of 100 natural population volunteers [7]. In 2015, the United States proposed the «Precision Medicine Initiative» (Precision Medicine Initiative) and prepared to launch the «All of US Research Program» with the goal of establishing a national cohort, measuring 1 million people and establishing a health database.
In 2003, British government officially launched the «UK Biobank Research Program». The project collects information on 500,000 volunteers from the UK’s National Health Service, including blood samples, genetics and lifestyle data, to explore the complex associations between genes and environmental exposures that affect common diseases [8]. In 2012, Imperial College London established the MRC-NIHR National Phenome Centre (NPC), relying on the relevant facilities of the London Olympics doping testing laboratory, to conduct related metabolic research through blood and urine. Its phenotype research is mainly based on different institutional centers scattered across the UK, focusing on disease and metabolic phenotype research.
In 2018, Germany invested in the establishment of the National Research Data Infrastructure (NFDI). In 2020, the European Molecular Biology Laboratory, the German Cancer Research Center, etc. established an alliance to officially launch the German Human Genome-Phenome Archive as part of the NFDI proposal, aiming to provide infrastructure and guarantee personal data security, etc.
In 2019, Murdoch University in Australia led the establishment of the Australian National Phenome Centre and set up a Western Australian Health Translation Network of several universities and hospitals. Based on the phenotype measurement platform, it takes the aim of studying diseases such as diabetes, cancer, and autism by measuring metabolic-related phenotypes.
In addition, Japan, Iceland, Luxembourg, Finland, France, Canada, Sweden, the Netherlands and other countries have also carried out phenotype research projects based on large population cohorts. International companies, including Google, are also actively conducting health-focused phenotypic research.
In general, following the Human Genome Project, developed countries, especially in the world, have successively carried out human phenotype research or large-scale health plans at the national or regional level, actively opening up a new era of human phenotype research. However, these phenotypic research efforts are limited to scattered phenotype measurement platforms, or focus on specific populations or types of phenotypes, lacking systematic, large-scale, standardized phenotype measurement platforms, phenotype group integration studies, and human phenotypes. In a relative sense, People’s Republic of China is the first country to systematically deploy human phenotype research. After years of construction and accumulation, it has developed cross-scale, multi-dimensional, and panoramic phenotype measurement and research work with an important contribution.
Phenotype groups: the «key» of a new era in life sciences
Why study phenotypes
The association between genes and phenotypes is very complex, and it is difficult to explain many complex diseases, such as diabetes and cancer, which are caused by environmental exposure and involve a large number of genes through genomics research. Phenotype studies are needed to delve deeper into the important functions of genes and their impact on health and disease. As gene sequencing and analysis technologies become more mature and affordable, the rate-limiting step in life science research has gradually shifted from gene sequencing to phenotypic measurement [4].
Through phenotypic measurements and massive amounts of genetic information, we can better compare the differences in traits between individuals. Based on phenotypic and epidemiological data from electronic medical records, phenotype studies on clinical diseases of community populations, we can explore the impact of genetic variation on phenotypes [9]. In the context of the current global COVID-19 pandemic,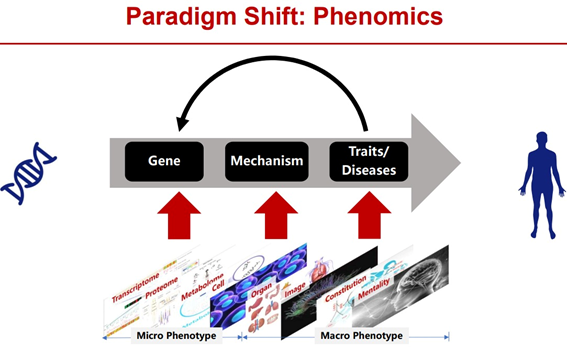 scientists suggest including phenotype and environmental studies to be one of the seven opportunities for achieving precision medicine [10]. Through phenotype research, it is beneficial to improve medical diagnosis and provide more reasonable disease treatment and prevention and control programs.
scientists suggest including phenotype and environmental studies to be one of the seven opportunities for achieving precision medicine [10]. Through phenotype research, it is beneficial to improve medical diagnosis and provide more reasonable disease treatment and prevention and control programs.
Relationships between phenotypes, phenotype groups and health
Phenotype studies involve measuring various multi-omics parameters, studying their impact on health and disease both at the individual and at population levels, and predicting treatment effects. Understanding the distinction between «normal», «healthy», and «disease» phenotypes, including defining the associations and transitions between these phenotypes, is very challenging for phenotype studies. For example, for an obese phenotype, a BMI (body mass index) greater than 30 kg/m2 is considered an abnormal range, but it does not necessarily lead to metabolic disease; for some natural populations considered to be normal. However, it may have health problems of metabolic obesity [11].
Through deep phenotyping and phenotype research, we can understand the dynamic characteristics of related phenotypes, detect important signal transitions and identify related molecular markers [12], which will help guide scientific personalized health management and Disease treatment interventions.
The research outcomes are in our service
During the transition from a healthy state to the state of disease, aging and environmental exposure often affect the living body. Through phenotype research, we will gradually realize the application of personalized health management and disease marker screening, based on big data of phenotype, and provide scientific and technological support and guarantee for the realization of Chinese and global health goals.
The team of Academician L. Hood from the Institute of Systems Biology in the United States studied aging-related diseases and health networks based on deep phenotype measurements, and combined multiple sets of biomarkers to provide lifestyle guidance [13]. The team of researchers Ding Ding and Cui Mei from Huashan Hospital of Fudan University and the team of researcher Chen Xingdong of the Institute of Human Phenotyping of Fudan University found that changes in metabolite levels are associated with the risk of Alzheimer’s disease, which is of great significance for early diagnosis and intervention of dementia [14] .
Based on the phenotype study of the Taizhou 泰州population cohort, this author’s team and others found that for five common tumors, including colorectal cancer, esophageal cancer, liver cancer, lung cancer, and gastric cancer, non-invasive blood testing can be performed earlier than routine diagnosis of early cancer screening. The screening of cancer biomarkers has important application value [15].
The team of Academician J. Nicholson of Murdoch University in Australia discussed the deep metabolic disturbance caused by the new coronavirus, and believed that phenotype research is very effective for measuring and monitoring the systemic pathological process of various diseases including new coronary pneumonia, Should be listed as an important tool for molecular studies and large-scale screening [16].
The importance of human phenotype research
Through human phenotype research, especially through the global collaborative human phenotype program, multiple results will be achieved in terms of scientific sources, precision health, and industrial cultivation.
In terms of scientific sources, by carrying out cross-scale and full-cycle precise measurement of human phenotypes on population samples, massive phenotype big data are obtained and analyzed, and large-scale discovery of gene-phenotype-environment and micro-phenotypes The cross-scale association and interaction with macro phenotypes draws a phenotype group reference map of related populations for understanding the mechanism of complex life processes. This will provide a new generation of «navigation maps» to explore strong associations between phenotypes and their related mechanisms, validate scientific hypotheses, and further enhance the original source of innovation in the field of life sciences.
The application and transformation of phenotype research and related scientific research results will provide lasting innovation power for the reform of the biomedical industry and the emerging growth points of the big health industry. Human phenotype research will discover a number of brand-new phenotypic markers, obtain new drug targets and new mechanisms on the whole scale, and provide new diagnostic reagents and products, next-generation original innovative drugs, personalized health management equipment, smart medical devices and a super engine for equipment, etc.; form a high-precision biological and health database, it provides a rich, solid and diverse data foundation for the cultivation and development of the big health industry based on «phenotype + big data» and «phenotype + artificial intelligence» .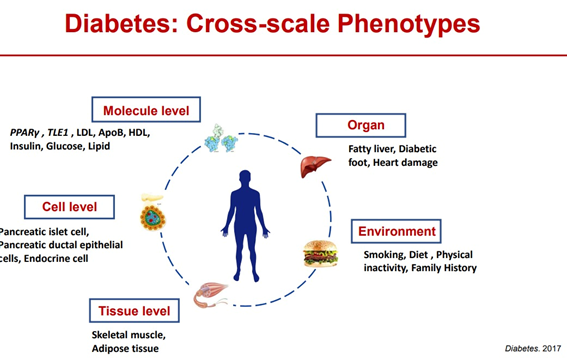
In addition, China has built the world’s first cross-scale, multi-dimensional human phenotype precision measurement platform and self-developed whole-process, automated phenotype big data platform, the country promotes phenotype measurement and analysis through phenotype research. It actively integrates into the global innovation network, promoting the development of personalized health management and diagnostic assistants, and leading the cross-generational development of the big health industry. Following the Human Genome Project, the International Science Project on Human Phenotypes, which has attracted much attention in the field of life sciences, will promote global technological innovation and progress in science, industry, society, etc. etc. are of fundamental significance.
Human Phenotype Study
Requires global collaboration and whole-of-society engagement
China takes the lead in deploying human phenotype research
China has diverse human genetic resources of 56 ethnic groups, which is of great significance for the establishment of representative population cohorts and phenotype studies. Human phenotype research in China started in the 1980s. Since 2014, Fudan University has been preparing to launch the International Major Science Program on Human Phenotypes. In 2015, the Ministry of Science and Technology supported the launch of the world’s first large-scale human phenotype research project «Investigation of Physical Human Phenotypic Characteristics of Various Ethnic Groups in China». In 2016, the Shanghai Municipal Science and Technology Commission gave priority support to the research on the phenotype group; in the same year, the State Council to list the «International Human Phenotype Group» as a major scientific basic project. In 2017, the first batch of major municipal science and technology projects in Shanghai, the «International Human Phenotyping Project (Phase I) Project», received approval. In order to lead the layout of the international major scientific plan of human phenotype, China has initially explored the «four-in-one» basic framework – platform construction, data system, collaborative network, and scientific research.At the same time, China has held a series of international academic seminars on human phenotypes, producing an international discourse and academic influence in the field of human phenotypes. In 2015, at the Xiangshan Science Conference with the theme of «International Human Phenotype Research», the participating experts at home and abroad reached a consensus and unanimously suggested launching the Human Phenotype Project. In 2016, the first International Human Phenotyping Symposium was held at Fudan University, advocating the launch of the International Human Phenotyping Research Program. In 2018, the 2nd International Human Phenome Symposium was held in Shanghai to define the implementation roadmap, cooperation mechanism and organizational structure of the Human Phenome International Science Program. HPCC (the Human Phenome Consortium of China) announced the establishment of a domestic collaborative network of human phenotypes co nsisting of 33 domestic universities and 22 top three hospitals; the author of this paper, J. Nicholson and L. Hood jointly proposed the establishment of the IHPC (International Human Phenome Consortium), mastering an international cooperation with 23 renowned research institutions in 18 countries. In 2019, Fudan University took the lead in organizing a new R&D institution, the Shanghai International Human Phenotype Research Institute, to promote the construction and standardization of the human phenotype R&D collaborative network at home and abroad. In 2020, through the online holding of the 3rd International Human Phenotyping Symposium, scientists from various countries in the Human Phenotyping International Collaborative Group reached a consensus on the three major directions that the Human Phenotyping Science Program that will place the focus, in the near future, on the fields including «COVID-19». “Phenotypes Research of Pneumonia and Other Major Diseases”, “Phenotype Research Technology System and Scientific Research Infrastructure Construction”, and “Standard Operating Procedures (SOPs) in Phenotype Research”. In 2021, the third IHPC Council, held online, adopted the principles for sharing phenotype data, and released the world’s first human phenotype navigation map, which pointed out the direction for the implementation of the human phenotype international big scientific plan.
of a domestic collaborative network of human phenotypes co nsisting of 33 domestic universities and 22 top three hospitals; the author of this paper, J. Nicholson and L. Hood jointly proposed the establishment of the IHPC (International Human Phenome Consortium), mastering an international cooperation with 23 renowned research institutions in 18 countries. In 2019, Fudan University took the lead in organizing a new R&D institution, the Shanghai International Human Phenotype Research Institute, to promote the construction and standardization of the human phenotype R&D collaborative network at home and abroad. In 2020, through the online holding of the 3rd International Human Phenotyping Symposium, scientists from various countries in the Human Phenotyping International Collaborative Group reached a consensus on the three major directions that the Human Phenotyping Science Program that will place the focus, in the near future, on the fields including «COVID-19». “Phenotypes Research of Pneumonia and Other Major Diseases”, “Phenotype Research Technology System and Scientific Research Infrastructure Construction”, and “Standard Operating Procedures (SOPs) in Phenotype Research”. In 2021, the third IHPC Council, held online, adopted the principles for sharing phenotype data, and released the world’s first human phenotype navigation map, which pointed out the direction for the implementation of the human phenotype international big scientific plan.
With the gradual development and optimization of the domestic and foreign collaborative innovation network of the Human Phenotype Project, the Chinese Human Phenotype Research Collaborative Group has gathered 77 members including 30 academicians; the Human Phenotype Project International Collaborative Group has gathered in Europe and the United States, the main research forces of major developed countries and countries along the “Belt and Road:” attracting 23 internationally renowned scientists, including 10 academicians from China, the United Kingdom, the United States, and Germany, to participate in the study of the human phenotype, which is a representative of the human phenotype. The global public technology platform and academic exchange center for group research has established a domestic and foreign cooperation mechanism.
Advancing human phenotype research requires multifaceted collaboration
Human health problems are global problems, not limited by countries, geography and other factors, especially for developing countries, the health threats caused by diseases are constantly rising. At present, bringing together global forces to solve common key problems in the field of human health through international major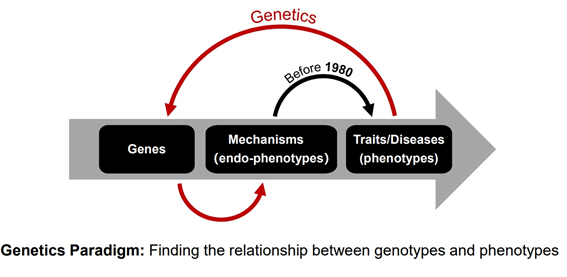 scientific programs has becom e the main scientific organization model. The Human Phenotype Project is a systematic, integrated and engineered project. It needs to be implemented through international scientific innovation cooperation at the level of platform construction and scientific research governance to promote the research of the human phenome.
scientific programs has becom e the main scientific organization model. The Human Phenotype Project is a systematic, integrated and engineered project. It needs to be implemented through international scientific innovation cooperation at the level of platform construction and scientific research governance to promote the research of the human phenome.
Germany, the United States, the United Kingdom, China, and Australia have successively established human phenotype research platforms. To make better use of the globally dispersed human phenotype platform resources, global collaboration in platform construction, phenotype measurement, and analysis standards is critical. The Human Phenotype Project Collaborative Group has set up a Technology and Standards Committee, which aims to unify the technical standards for phenotype measurement and analysis of the platform, ensure the authenticity, accuracy, and comparability of data, and maximize the use of existing platforms, projects and other resources to provide human phenotypes. Group research provides the core foundation.
Human phenotype research involves multidisciplinary fields such as biology, medicine, data science, ethics, public affairs, information sharing, and security. The participating countries and organizations are diverse and complex, and it is necessary to improve the management structure and operation mechanism. Including decision-making consultation, supervision and evaluation, risk prevention and control, etc., implement resource and achievement sharing, complementary advantages, and risk sharing, and comprehensively represent human beings in terms of funding, intellectual property, social ethics, data security, talent gathering and training, etc. The implementation of the type group plan is escorted.
Human phenotype research needs
the support and participation of the society
Human phenotype research takes individuals or specific groups as the research  object, and it requires the support and participation by the public, scientific community, industry and other forces of the whole society, to carry out the planning, construction and operation of the human phenotype project in an orderly manner.
object, and it requires the support and participation by the public, scientific community, industry and other forces of the whole society, to carry out the planning, construction and operation of the human phenotype project in an orderly manner.
Human phenotype research requires the participation and support of the public as test volunteers. Longitudinal in-depth phenotype measurements at the individual level are of great significance for scientific research and personal health management from healthy condition to state of disease; cohorts based on specific populations Research reveals statistical distribution and influencing factors from a group perspective, and provides scientific evidence for individualized treatment and precise prevention. Human phenotype research also involves related social issues such as ethics, law, and privacy, and requires public participation, supervision and joint improvement.
In addition, human phenotype research requires the continuous participation and linkage of the scientific community, business community, etc. Human phenotype research will provide a source of innovation for scientific research and the health industry, and promote the coordinated development of basic theoretical research, new diagnostic reagents, original innovative drugs, new technologies and equipment.
Conclusion
With the advent of the post-genome era, the significance of phenotype research has become increasingly significant. Through preparation and launching of the Human Phenotype Project, we will break through the bottlenecks in the field of life sciences and medical innovation, comprehensively interpret the code of human life and health, and lead the exploration of the human microcosm. The future journey will help realize the beautiful vision of a community of human health and health.
(Translated by Jiger Janabel from Science-Bimonthly Journal of China. No 28.012022)
Toward the Depth of Phenotypic Understanding
— a preliminary report on recent findings from the sample collection
Gulnur Junusova, Anastasiya Perfilyeva, Kira Bespalova, Nazym Altynova, Azada Khamdieva, Djansugurova Leyla
The Institute of Genetics and Physiology (IGP), in collaboration with the Institute of Oncology, is conducting a project in cancer related issues through the means of multi-gene sequencing technology. This allows us to identify pathogenic mutations in different genes. In our studies, we have identified mutations that are characteristic of a particular type of tumor, as well as new mutations that were previously unknown. Some mutations have been found to be associated with early development of the disease as well as with certain syndromes and phenotypes or with an unfavorable prognosis of the disease. The Molecular Genetics Laboratory of the IGP has been conducting research on various cancers: breast, prostate, cervical, esophageal, and colorectal. These studies have identified specific genetic markers associated with the risk of developing these cancers and with diagnostic and prognostic biomarkers. Based on the results of NGS sequencing in patients with colorectal cancer, we have been able to identify the potentially pathogenic variants, including novel variants not previously registered. These variants were evaluated for functional impact and prognostic value (high and intermediate penetrance genes listed in the National Comprehensive Cancer Network NCCN guidelines).
The frequency of pathogenic mutations in CRC genes with high penetrance was higher in patients with primary multiple cancers (21.1% vs. 3.1%; p = 0.0002) and in patients with FAP syndrome (20.0% vs. 3.1%; p = 0.0004) than in patients without a family history of cancer (3.1%). Based on the results of studies of patients with breast cancer, a panel of genetic markers for hereditary predisposition to breast cancer was established, including 16 mutations in 8 genes. In addition to mutations common in breast cancer, mutations characteristic of female Kazakh group who are identified for the first time (founder mutations in BRCA1 and BRCA2). These mutations are characterized by early development of the disease. Identification of genetic markers associated with the development of certain types of cancer in the Kazakh population will allow to reach a new level of research and increase the efficiency of identification of cancer patients at an early stage of the disease. Knowledge of the mutation spectrum will enable more efficient updating of screening protocols and allow the development of a faster, more accessible and less costly strategy for genetic testing.
than in patients without a family history of cancer (3.1%). Based on the results of studies of patients with breast cancer, a panel of genetic markers for hereditary predisposition to breast cancer was established, including 16 mutations in 8 genes. In addition to mutations common in breast cancer, mutations characteristic of female Kazakh group who are identified for the first time (founder mutations in BRCA1 and BRCA2). These mutations are characterized by early development of the disease. Identification of genetic markers associated with the development of certain types of cancer in the Kazakh population will allow to reach a new level of research and increase the efficiency of identification of cancer patients at an early stage of the disease. Knowledge of the mutation spectrum will enable more efficient updating of screening protocols and allow the development of a faster, more accessible and less costly strategy for genetic testing.
Advanced panels of genetic markers can be used by medical institutions and clinical diagnostic centers to screen high-risk groups (patients and their close relatives). Screening results can be used for timely medical genetic counseling of high-risk patients. Information about the specific mutations that caused the disease will allow timely preventive measures to be taken and the best treatment tactics to be selected. This approach is the introduction of the principles of personalized medicine into practice and will have a tangible social and economic impact. In collaboration with medical institutions, the Institute conducts active research in the field of molecular genetics of multifactorial diseases. Gene banks have been established for patients with cardiovascular, oncological, neurological and allergic diseases (about 1,500 individuals in total).
Autism Spectrum Disorders and Epilepsy
Autism Spectrum Disorders (ASDs) are heterogeneous syndromes defined by impairments in three key areas: social interaction, speech, and interest domain. Our studies at IGP show that heterogeneity, both genetic and phenotypic, is the dominant feature of ASDs. Given this heterogeneity, a better understanding of the genetic factors underlying ASD requires many parallel approaches to the research process. These methods include the collection of large, well-characterized cohorts of patients and their relatives representing not only different phenotypes but also different populations, genome-wide studies, studies involving a joint consideration of multiple etiologic factors, and association studies. In 2018, the Institute of Genetics and Physiology launched a project to study the molecular genetic aspects of autism in Kazakhstani populations. In the course of reaching the goals, we created unique databases with personal data and biological material collected from 400 families in Kazakhstan where children were diagnosed with ASD. Using database mentioned above, the team has investigated polymorphisms in a number of candidate genes involved in the metabolism of biologically active substances, receptor genes, etc. As a result, polymorphic markers have been identified that are associated with an increased risk of developing ASD in a child or with more severe manifestations. In the presence of a high degree of loci heterogeneity (as is very likely in ASD), a potentially useful approach to identifying candidate variants is to examine large families with many affected individuals.
Examining the broadly defined ASD-related traits, such as BAP (broad autism phenotype) is another possible way to increase the power to detect candidate variants. We applied these methods in our study, which is one of the first genomic studies of families with a hereditary problem of autism in Kazakhstan. The results obtained allow us to discuss the relative merits of searching for genetic causes of the disease in large pedigrees related to neuropsychiatric disorders and also clearly show how many genes and metabolic pathways underlie the development of ASD. We studied a total of 4 families (19 individuals, including 8 children with ASD) with hereditary ASD burden. Using genome scans on the Illumina® Global Screening Array (GSA) platform, we identified dbSNPs associated with ASD development, as well as rare inherited variants and de novo mutations. When we examined de novo gene variations, we found that affected siblings carried discordant mutations associated with nervous system disorders while exhibiting varying degrees of ASD severity. These data suggest that the identified dbSNPs and rare heritable variants exerted a general genomic burden and determined the inherited form of ASD, whereas de novo mutations determined the clinical severity of autism.
to increase the power to detect candidate variants. We applied these methods in our study, which is one of the first genomic studies of families with a hereditary problem of autism in Kazakhstan. The results obtained allow us to discuss the relative merits of searching for genetic causes of the disease in large pedigrees related to neuropsychiatric disorders and also clearly show how many genes and metabolic pathways underlie the development of ASD. We studied a total of 4 families (19 individuals, including 8 children with ASD) with hereditary ASD burden. Using genome scans on the Illumina® Global Screening Array (GSA) platform, we identified dbSNPs associated with ASD development, as well as rare inherited variants and de novo mutations. When we examined de novo gene variations, we found that affected siblings carried discordant mutations associated with nervous system disorders while exhibiting varying degrees of ASD severity. These data suggest that the identified dbSNPs and rare heritable variants exerted a general genomic burden and determined the inherited form of ASD, whereas de novo mutations determined the clinical severity of autism.
Although we could not confirm with certainty whether any of the identified genetic variants actually contribute to the disease, our analysis provides a list of priorities for further validation studies. The most interesting aspect of the study is that all mothers from four families had biallelic mutations in the ACADS gene, which plays an important role in mitochondrial oxidation of shortchain fatty acids. We hypothesize that maternal and fetal energy deficits caused by the mutations in the ACADS gene may contribute to the development of ASD under stressful conditions. Primary ASD-associated changes occur during intrauterine development of the fetus and are manifested/increased in the child during the postnatal period when exposed to environmental factors. These and other data lead us to conclude that a disease as complex as ASD should be studied in conjunction with other factors. The presence of a well-characterized database, the support and agreement of the parents of children with ASD to further collaboration with us, gives us hope for the success of further in depth studies, which will include not only the study of genomes, but also the analysis of biochemical parameters, physiological and psychological studies.
Epilepsy is a chronic neurological disease with a high degree of genetic heterogeneity, which is accompanied by recurrent unprovoked seizures and with various clinical manifestations. Genetic factors (70-80% of cases are of a genetic nature) are one of the main causes of epilepsy, but search these factors is difficult due to the extreme heterogeneity of the disease. Previously our institute (in 2018) carried out a research project where we studied the molecular genetic aspects of hereditary forms of epilepsy in Kazakhstani populations (AP05131747). Within the project we created a database of personal data of epilepsy patients, we collected biological material and DNA of 154 patients with various forms of epilepsy and 154 apparently healthy individuals, and performed a molecular genetic analysis of the candidate genes of epilepsy focused on the clinical diagnosis of patients. We performed a molecular genetic analysis of 24 genetic variants and polymorphisms in the ion channel genes (SCN1A, SCN2A KCNT1, KCNC1, KCNQ2), in the γ2 gene of the gammaaminobutyric acid receptor subunit (GABRG2) and in the gene of the glucose transporters (SLC2A1). The results obtained give grounds to consider that SCN1A, KCNT1 and GABRG2 have a definite pathogenetic significance in various forms of epilepsy with generalized and focal seizures. For a single nucleotide variant of the T allele of the SLC2A1 gene, an associative risk of developing epilepsy was identified. The carriers of the SLC2A1 458Trp allele were patients with various forms of epilepsy, manifested by generalized, but not focal seizures. Our results indicate the possibility of detecting pathological mutations in the SCN1A, KCNT1, and GABRG2 genes even in a small group of patients with non-mechanical forms of epilepsy. We also performed sequencing of whole mitochondrial DNA of patients who had maternal inheritance. We performed genome-wide SNP genotyping for patients with unknown etiology of epilepsy and resistance to anticonvulsants. According to the genome-wide analysis, 21 variants associated with epilepsy were identified. Allergy is one of the diseases of peacetime and a kind of marker for the well-being of the country. WHO has been seriously concerned for 25 years about the epidemic spread of allergies, as well as the fact that among all the main causes of death, mortality from respiratory diseases continues to increase and systematically comes to the fore. For this type of diseases, IGP has collected blood samples from patients with various allergic diseases. According to the category of types of allergic diseases, the following groups of patients with diagnoses were distinguished based on the data of allergic history of physicians: allergic rhinitis, atopic dermatitis, urticaria, drug allergy, acute bronchitis, bronchial asthma, toxic erythema, Quincke’s edema, allergic conjunctivitis, solar urticaria, food allergy.
To analyze new and old genebank material, genome wide analysis based on next generation sequencing (NGS) and microarray genotyping and associated bioinformatics analysis technologies we test the connections between hundreds of thousands of single nucleotide polymorphisms and various effects and characteristics of disease. Such associative analysis will enable us to determinate the genetic risks in individuals exposed to mutagenic environmental factors, identification of causal mutations in patient cohorts with allergies, epilepsy, autism, and prostate cancer, provide more precise answers to mechanisms of disease development, and suggest diagnostic interventions.
Within the framework of the Population Genetics Laboratory (established in 2014), unique in Kazakhstan, which focuses on interdisciplinary research and strengthening the scientific and evidential base of Kazakhstan’s national history using modern capabilities of molecular genetic analysis, Kazakh geneticists have built a genebank that stores genes from as many as 2000 individuals of different genealogical background. For many years, the Institute of Genetics and Physiology has been studying the effects of radiation and other mutagenic environmental factors (petroleum products and derivatives, rocket fuel, pesticides) on the genome of humans, animals, and plants from ecologically harmed regions of the Republic of Kazakhstan (Semipalatinsk Test Site, Caspian Sea, Aral Sea, Ile Balkhash, Almaty Region, etc.) and as a result of occupational damage.
A priority of the Institute is the study of the long-term consequences of the impact of technogenic factors on the human genome. Gene banks representing human populations from ecologically unfavorable regions (more than 1000 samples) and corresponding control populations from ecologically favorable regions (more than 500 individuals) have been collected. Technology has been developed to allow comprehensive investigation of the effects of anthropogenic environmental factors on the human body at the gene, chromosome, and population levels. In the future, we plan to move to exome or whole-genome sequencing, which can be used to study a number of multifactorial, socially significant diseases with hereditary predisposition, such as cardiovascular diseases, allergic diseases, autism, etc.

